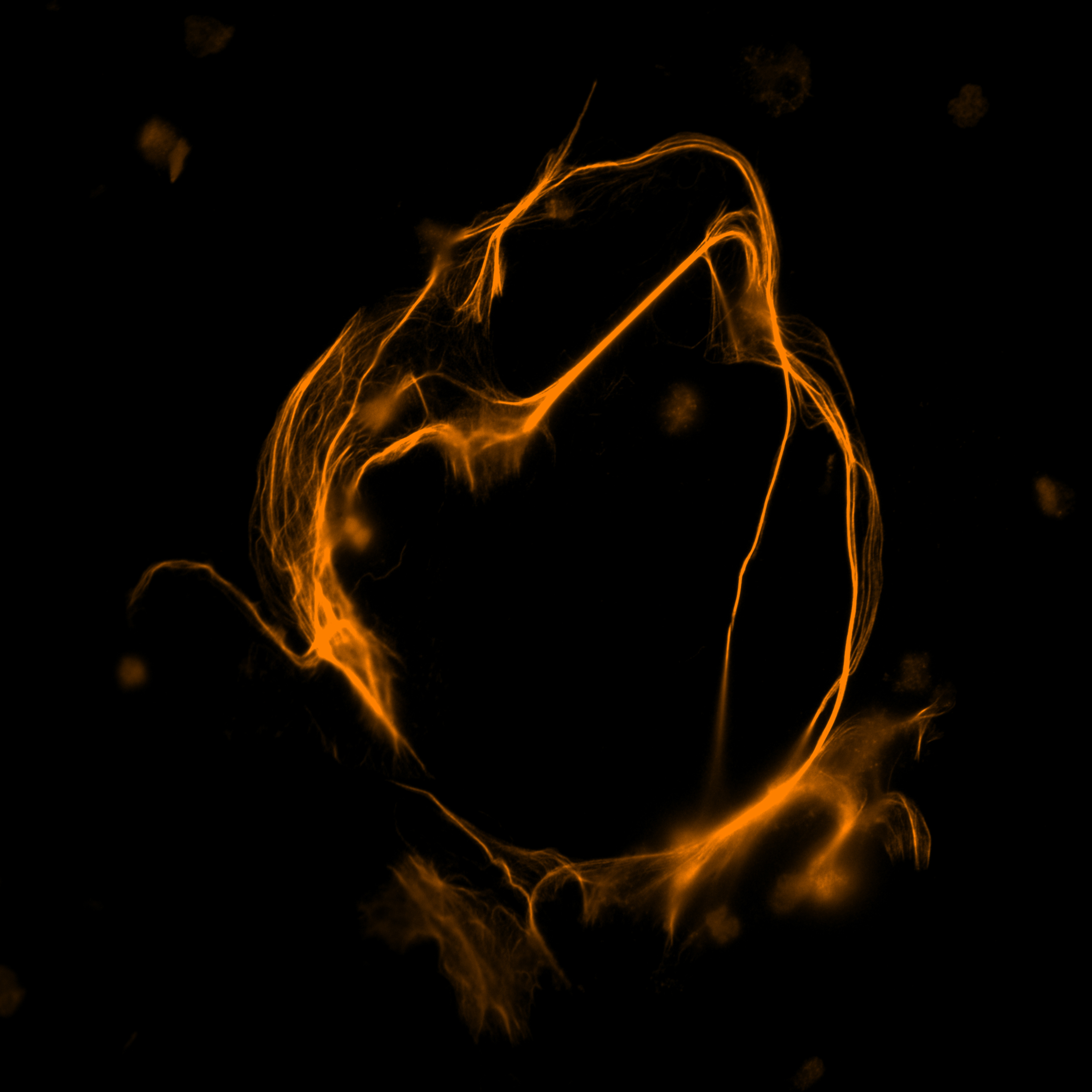
Top Prize Image in Visualizing Science Contest Captures Research Tied to the Sun
Ph.D. student Maile Marriott’s submission illustrates the complexities of the “space weather” generated by our sun.
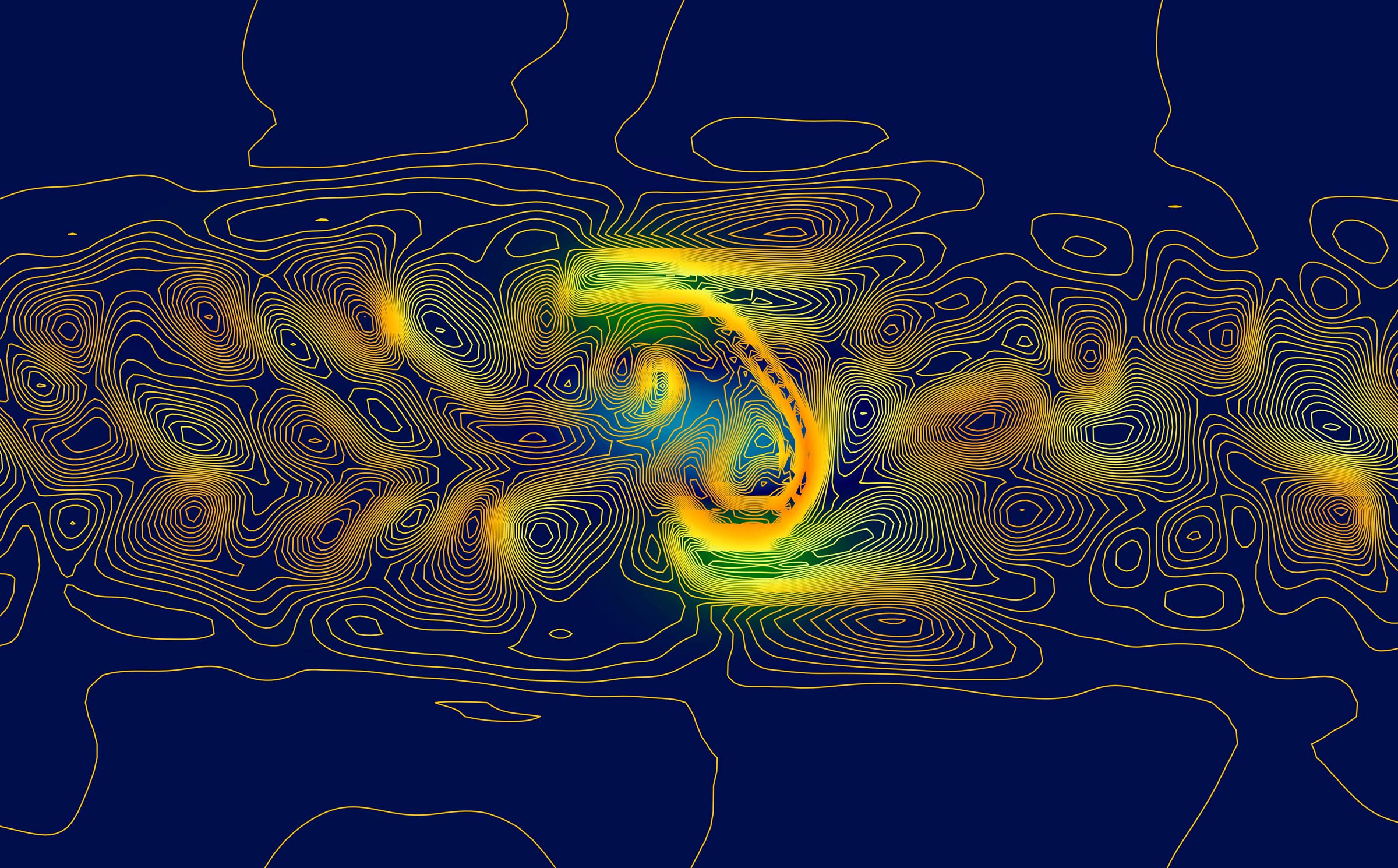
Top Prize Editor’s Choice by Maile Marriott, Physics Graduate Program
Each year, the College of Natural Sciences invites University of Texas at Austin faculty, staff and students to send in the best images from their research for our Visualizing Science competition. The wondrous images that result often simultaneously inform the mind and warm the soul.
Consider physics graduate student Maile Marriott’s submission, which received top honors and illustrates the complexities of the “space weather” generated by our sun. Researchers like Marriott are looking forward to the total eclipse that is set to pass over Texas and many parts of North America on April 8. One reason is that this year’s eclipse also coincides with what is known solar maximum, a period of heightened solar activity that happens only every few years.
“The eclipse will give us a great view of the corona and events happening closer to the sun’s surface,” Marriott added, “things that normally are hard to view because the sun’s light is too bright at other times.”
Because scientific imagery has the power to inspire – and a perfectly-timed photo or stunning visualization can draw the viewer in and encourage curiosity in new areas of interest – past winners of Visualizing Science are currently on display at the Texas Science and Natural History Museum on the UT Austin campus.
For 2023-2024, eight outstanding submissions from the Natural Sciences community were chosen as finalists for their beauty and scientific merit, selected by professional science communicators or by community members on social media.
The winning entries on this page can also be experienced in our Visualizing Science Showcase, a 3D virtual gallery exhibition featuring winners and finalists from more than a decade of competitions. Select images in the gallery include expanded audio descriptions by their creators. Wander through the virtual gallery using the link at the bottom of this page.
About the Top Prize Editor’s Choice
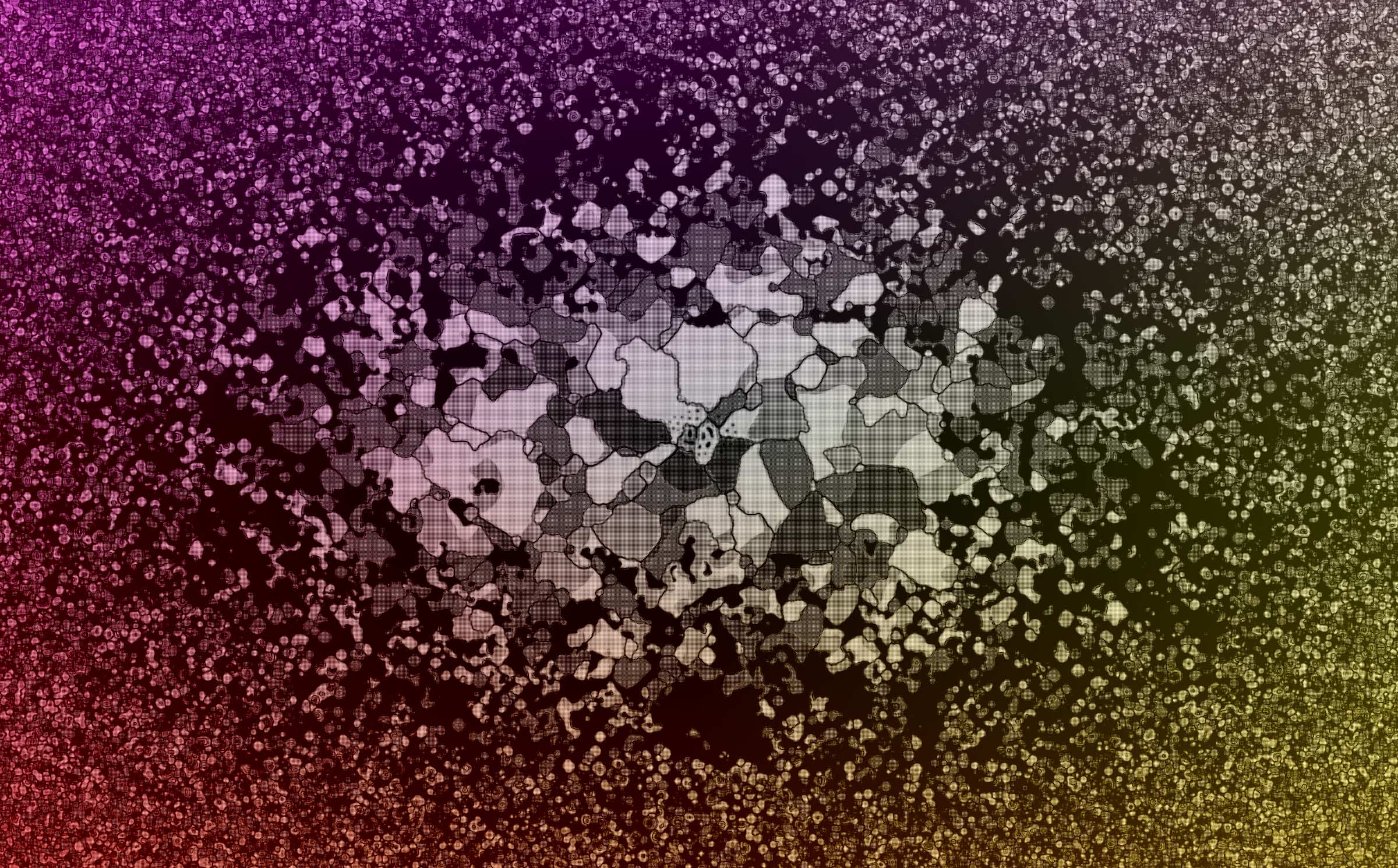
The solar wind blows outward from the sun, generating what we might think of as space weather that affects our satellites, space missions and even our magnetosphere, atmosphere and life here on Earth. Switchbacks are a common but little-understood phenomenon in the solar wind where the magnetic field lines reverse direction and form a “kink.” Since the solar wind is a plasma, the flow of charged particles follows the kinked path of the magnetic field, like water following the wave of energy as it moves towards the seashore.
This image is generated from a Magnetohydrodynamic (MHD) simulation of a switchback. Light blue is where the magnetic field is negative (reversed direction), while green is where the field is positive. The gold lines show the plasma density, and the lines swirl tightly around the kinked form of the magnetic field.
The purpose of this simulation is to study how the switchback decays and what factors make it more stable in the solar wind, so that researchers can better understand how these structures are made and how they might contribute to turbulence and heating in the solar wind.
Additional 2023–24 Winners
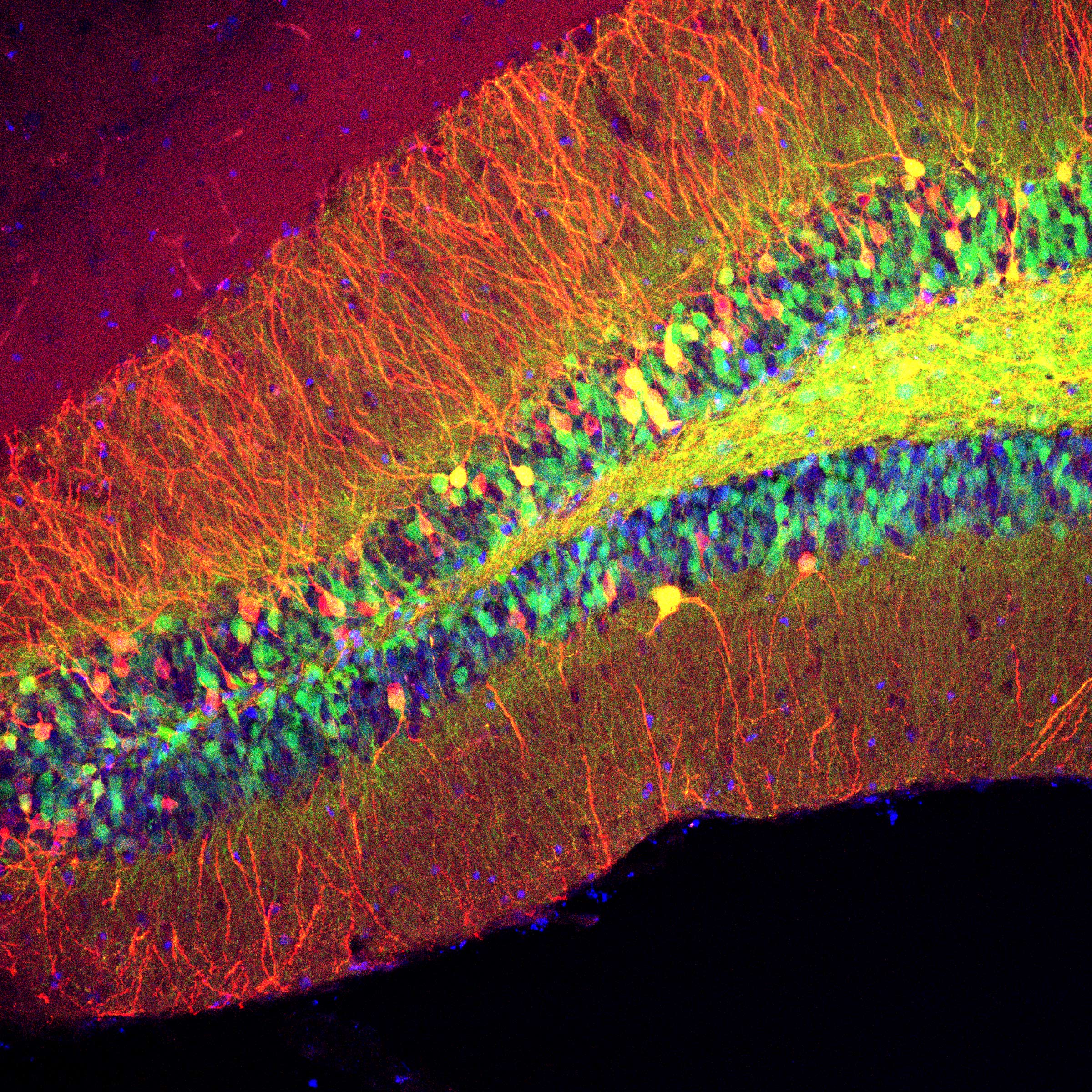
Nathaniel Nocera (Ph.D., ’21), Neuroscience
The hippocampus is an evolutionarily conserved brain region that is essential for learning and memory in mammals. Memories of people, places, things, as well as our individual life experiences, are all encoded and recalled by the hippocampus. The Drew and Zemelman labs use viruses to visualize and alter cellular activity to understand how cells interact with one another both within and across regions of the brain during learning processes. By combining anatomical studies with behavioral experiments, researchers can search for patterns of activity in cells and regions and better understand how human memory works. Ultimately, the hope is to apply this knowledge to treat neurological disorders such as Alzheimer’s Disease.
In this image, granule cells in the dentate gyrus of a mouse hippocampus glow with fluorescent proteins. The hair-like filaments labeled in red (dendrites) transmit information to the multi-colored cell bodies. The diameter of these granule cells is ~15 microns, or a third the width of a strand of fine human hair.
Weijie Guo, Biochemistry Graduate Program
The human immune system deploys a vital defense mechanism that is captured in part in this image. Neutrophils, a type of white blood cell, release intricate DNA structures to ensnare and neutralize invading pathogens. This captivating confocal image unveils the intricate world of those structures, called neutrophil extracellular traps (NETs). Stained with a nucleic acid dye, NETs start to resemble a mesmerizing dance of chromatic threads. The vivid fluorescence captures the dynamic nature of this immune response, showcasing a beauty that lies within the frontlines of our body’s defense.
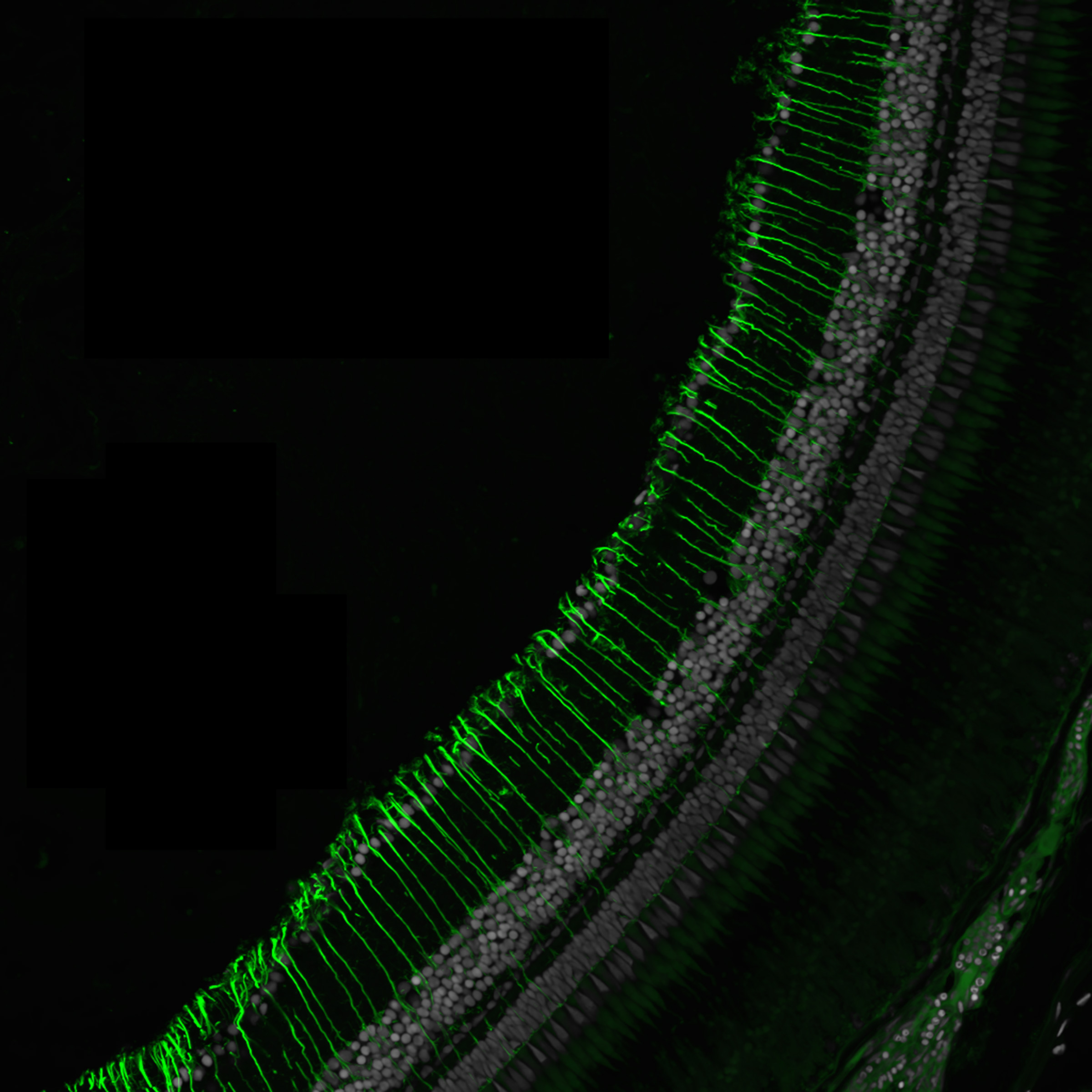
Reza Raeisossadati, Molecular Biosciences
To understand a range of vision-related disorders and opportunities for new therapeutic solutions to those, researchers in the lab of Jeff Gross study the eyes of zebrafish. Seen here is the retina of an adult zebrafish from a transgenic line that expresses a green fluorescent protein in cells known as Müller cells. Müller cells are the stem cells of the eye and can divide and regenerate into any cell type in the retina to repair damage. They become activated upon injury in zebrafish. The Gross lab uses this transgenic fishline to study and better understand regeneration in the eye.
Marco Ravelo, Computer Science Graduate Program
Some images and areas of study manage to represent a diverse cross-section of disciplines: computer science, mathematics, physics, biology and myriad others. Cellular automata, popularized by Conway’s Game of Life in the 1970s, is one such example.
Shown here is a snapshot of a version classified as a continuous cellular automaton, where a cell’s state can be represented not just by a discrete value (usually an integer), but by any rational number (in this case between 0 and 1). Each pixel in the image represents a single cell, and the color represents the cell’s current state. Every round, each cell undergoes a calculation in which it perceives the states of its eight immediate neighbors and chooses a new value for itself based on parameters defined by the programmer. When tuned correctly, these constant changes showcase a beautiful animation emblematic of the phenomena of emergence in mathematics and science.
Many of these automata display life-like characteristics and can grow incredibly complex. The program used to create this automaton was written by graduate student Marco Ravelo and can be accessed here as an interactive demo.
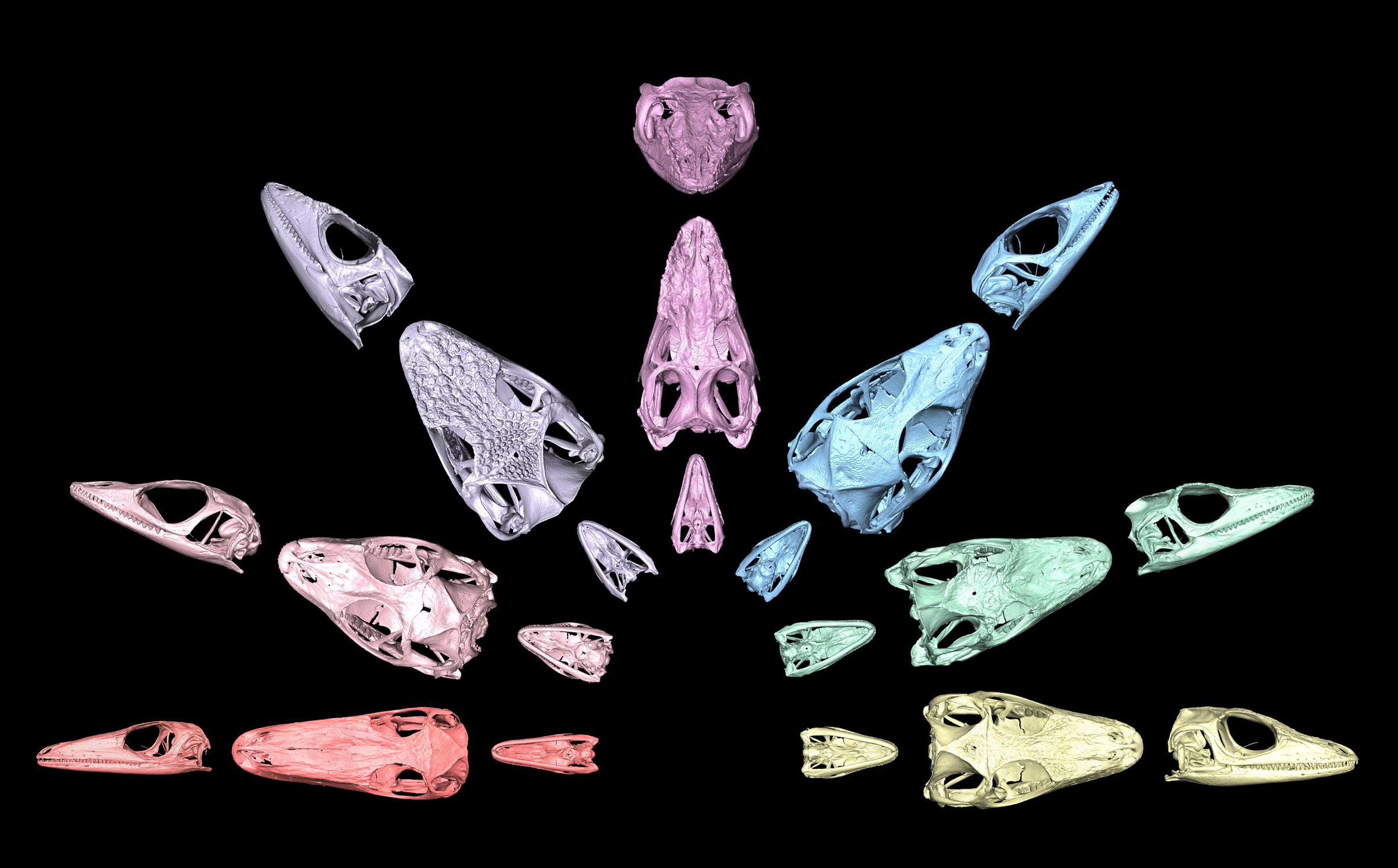
Tianyi Xu, Ecology, Evolution and Behavior Graduate Program
Anoles are a group of highly diverse lizards, having undergone the evolutionary process of adaptive radiation on different Caribbean islands. Across the Greater Antilles, anoles evolved distinct forms known as ecomorphs, which are local adaptations to different microhabitats and niches through convergent evolution. These different forms have very distinct sizes and shapes. These 3D, high-resolution X-ray computed tomography images of anole skulls showcase the variation. The lizards represented are, from left to right: grass bush, trunk, crown giant, trunk crown, trunk ground, twig and a unique semiaquatic variety. The skulls in the image are not to scale and were resized to similar dimensions for comparison purposes. Specimens were loaned from collections at the University of Kansas, American Museum of Natural History and Smithsonian National Museum of Natural History and scanned at UT’s High-Resolution X-ray Computed Tomography Facility.
Facebook Favorites
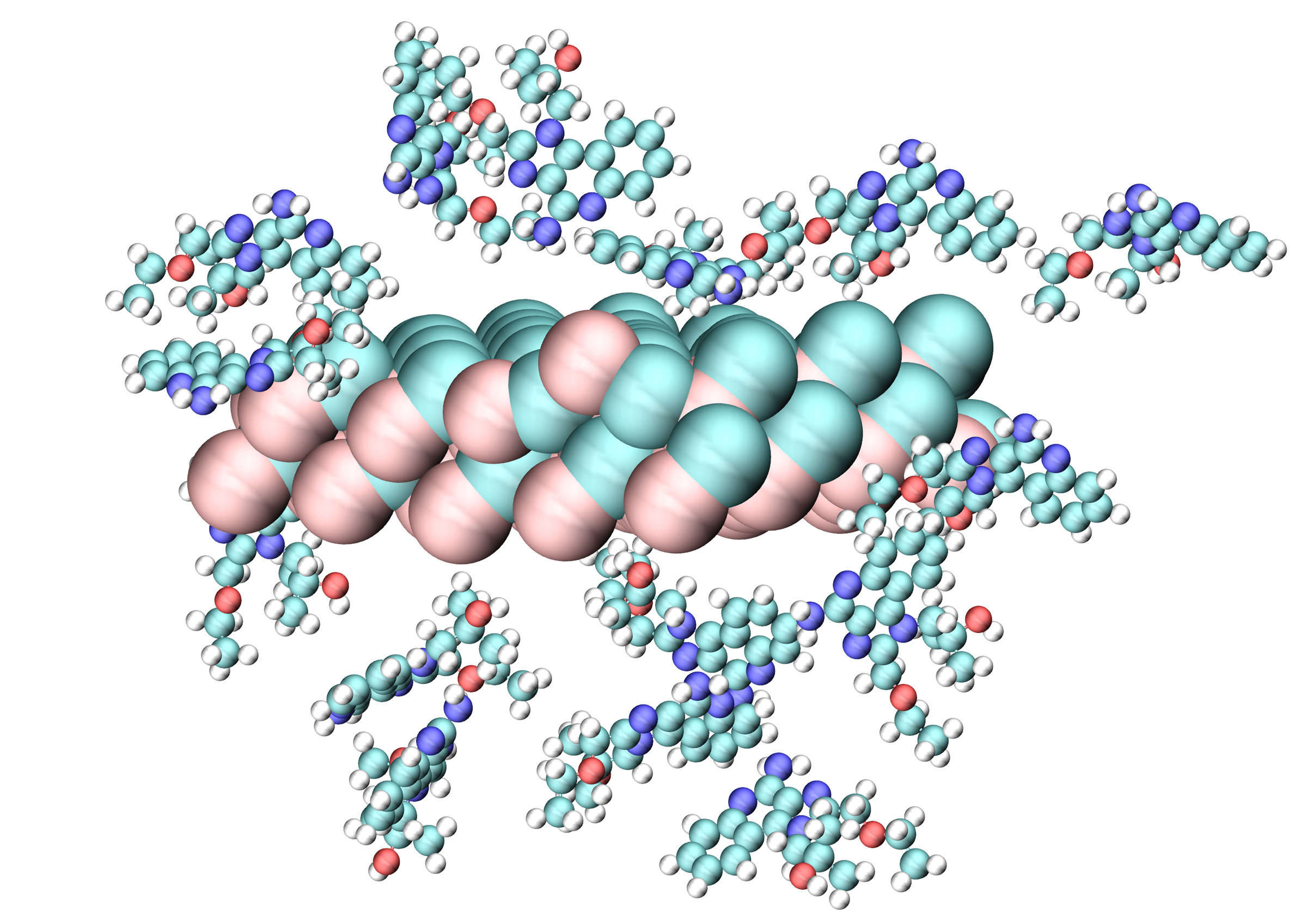
Mohammed Bhia, Chemistry Graduate Program
Computer-aided approaches can hold promise for rapidly screening many different options for a precisely engineered cancer therapy. For example, the environment surrounding cancer cells is acidic, while healthy tissues are neutral in nature. A visualization like this one demonstrates this contrast in acidity and can be used to show a plan to specifically target cancer, while safeguarding healthy tissue from drug side effects. The researcher’s goal is to release a special drug (resiquimod) to boost the immune system without harming healthy cells. To achieve this, Mohammed Bhia used computer simulations to simulate tiny inorganic particles known as mesoporous silica nanoparticles that can hold onto the drug in a neutral, non-acidic environment. These particles act like carriers, ensuring the drug is localized and does not affect healthy tissue by only releasing it in the acidic environment created by cancers. In this visualization, the simulations displayed reliable stability and drug containment in the near-neutral pH associated with healthy cells.
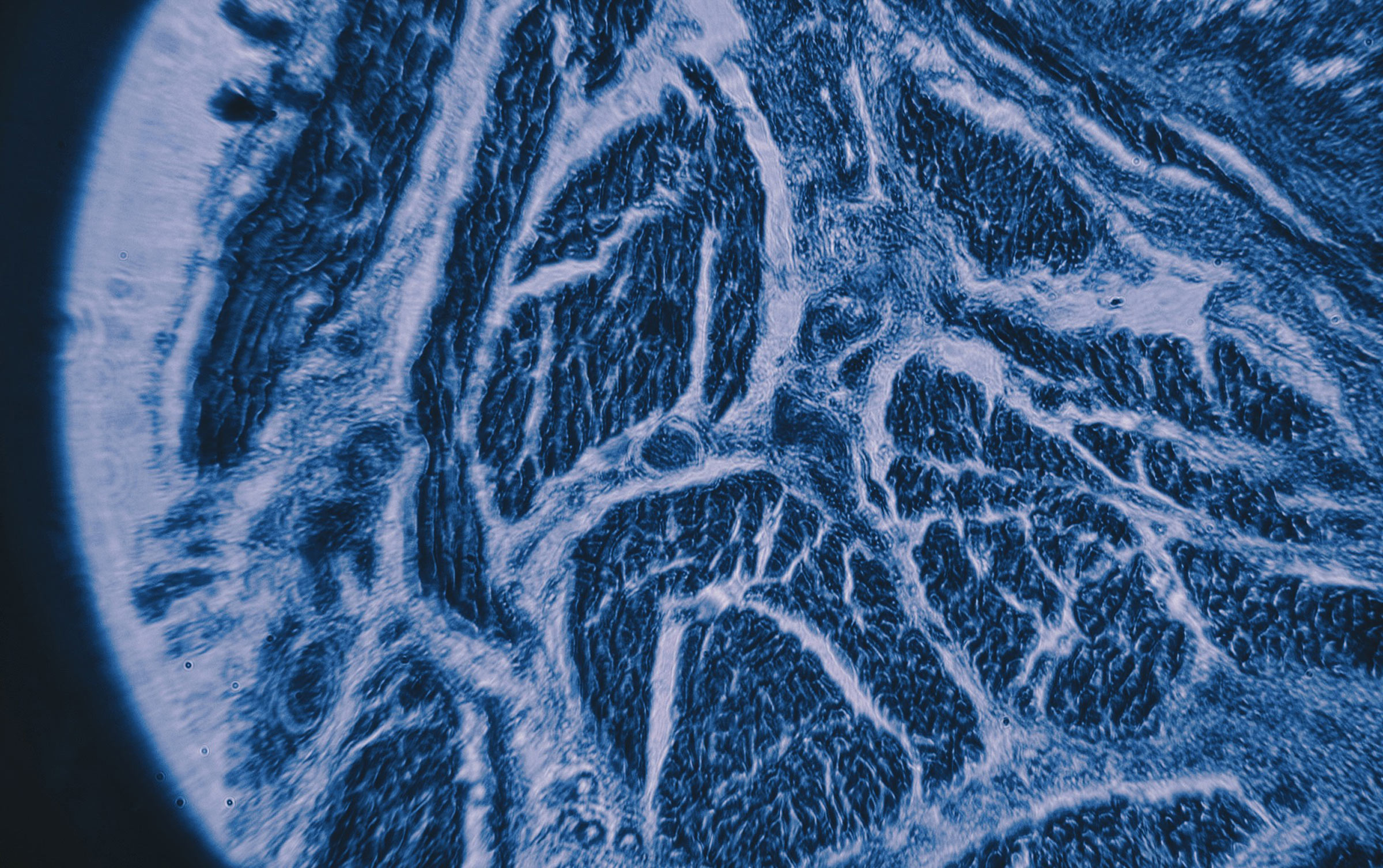
Alexa Cumming (B.S., ’23), Neuroscience
Alongside remarkable research to advance scientific discovery, the College of Natural Sciences is home to eye-opening and multifaceted educational experiences. In the image above, a human tonsil was captured through a light microscope for a neuroimaging laboratory course in the Department of Neuroscience (NEU 366L). Students in the course are tasked with putting together their own light microscope in an attempt to get the clearest magnification possible.
Visualizing Science Showcase